
Miguel Aréjula Aísa
- 21/07/2025
Clinical Pathway Optimizer: Improving Hospital Protocols with Formal Methods
A web-based system to digitize, monitor, and validate clinical pathways in real hospitals, developed as part of my Computer Engineering Bachelor's Thesis.
In the middle of healthcare’s digital transformation, many hospitals still rely on spreadsheets or manual notes to manage complex clinical processes. During my final year as a Computer Engineering student at the University of Zaragoza, I had the opportunity to work on a project that aims to improve this situation by introducing formal methods into real-world medical workflows.
What Is Clinical Pathway Optimizer?
Clinical Pathway Optimizer is a web-based platform I designed and developed as part of my bachelor’s thesis. It helps hospital staff to digitize, monitor, and improve clinical pathways, that is, standardized sequences of medical actions during a procedure such as a hip or knee replacement.
The system was created in collaboration with the Hospital Clínico Universitario Lozano Blesa and is aligned with a national research project focused on applying formal methods and process mining to real clinical data.
The Problem
Despite advances in hospital technology, many departments still track treatments manually. This leads to:
- Limited visibility into what was actually done during a patient’s operation.
- No objective way to verify if protocols were followed.
- Difficulty identifying what parts of a clinical pathway are inefficient or outdated.
Previous Work
Before I started building the web platform, part of the groundwork for this project had already been laid in the form of a Python-based algorithm developed in earlier research stages. This algorithm was capable of automatically generating clinical pathways from structured CSV data representing real-world medical procedures.
Its goal was to identify common patterns in clinical workflows and formalize them into Petri nets, graphical models commonly used in systems analysis to represent sequences of actions and their dependencies.
However, while the algorithm was technically sound, it had some major limitations:
It required a technical environment to run (command-line interface, Python dependencies). It lacked an intuitive interface for healthcare professionals. Its output was not easily interpretable or verifiable without specific expertise in formal methods. This is where my work began. Rather than reinventing the algorithm, I focused on integrating it into a broader system that could make it usable and useful in a real clinical setting.
The Solution
The main objective of this project was to bridge the gap between technical tools and clinical practice by building a complete system that:
- Enables digitization and structuring of clinical pathways using real data.
- Supports real-time monitoring of patient adherence to predefined medical protocols.
- Provides automatic validation of these protocols through formal analysis techniques like Petri nets.
- Makes the system usable by healthcare professionals without requiring technical expertise.
With these goals in mind, I designed and implemented a modular, web-based application that integrates the previously developed Python algorithm and exposes it through a user-friendly interface. The result is a system that allows hospital staff to both manage and continuously improve their clinical workflows.
Core Functionalities
-
Doctors can:
- Register new patients and surgeries.
- Track the progress of a patient through a clinical pathway by logging completed tasks.
- Search patients by medical record number and view treatment histories.
-
Admin-doctors can:
- Import clinical pathway definitions from CSV files.
- Automatically generate Petri nets from those definitions using the integrated algorithm.
- Validate the resulting models to ensure logical correctness (no deadlocks, guaranteed progress).
- Assign tasks to phases, define expected response types, and edit existing pathways.
Methodology
The development process followed an iterative and incremental approach, heavily influenced by direct feedback from clinicians. I conducted multiple requirement-gathering sessions with orthopedic surgeons at the Hospital Clínico Universitario Lozano Blesa to understand their workflows, constraints, and needs.
The project was structured into five main phases:
- Requirements analysis, based on real-world use cases from the hospital.
- Architecture design, with a focus on modularity and future scalability.
- Implementation, prioritizing features with immediate clinical value.
- Validation, including integration testing and automated model verification.
This close collaboration with medical staff ensured that the tool evolved in alignment with real hospital practices, not just technical assumptions.
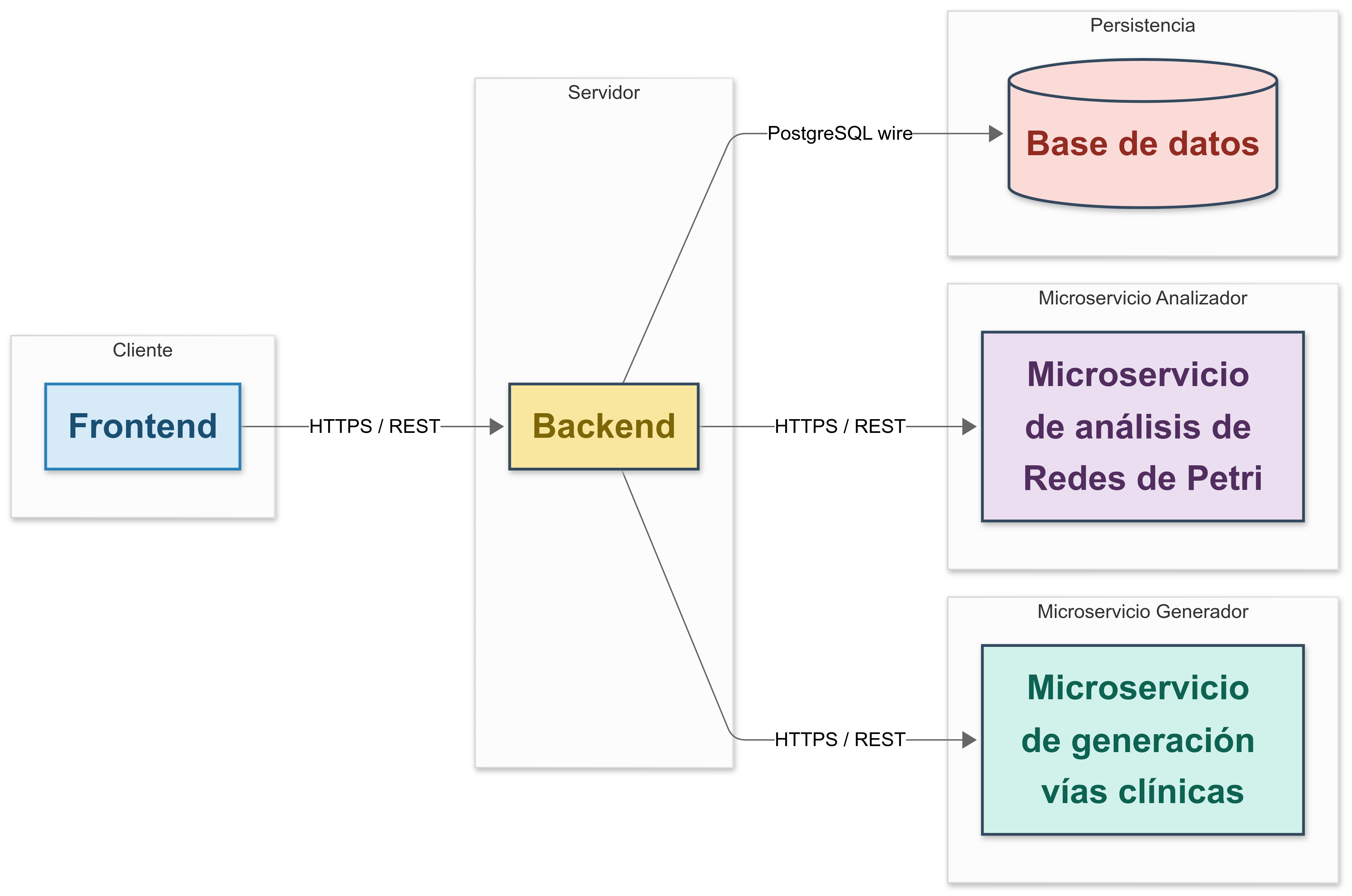
System Architecture
The application is built around a modular client-server architecture with support for microservices. This allows independent evolution of each component and simplifies future maintenance.
The system is composed of:
-
A frontend, built with React and styled using Tailwind CSS.
-
A backend server, developed with Node.js and Express, acting as the main API and coordinator.
-
Two Python microservices, built with FastAPI:
- One to generate Petri nets from CSV-based pathway definitions.
- Another to validate those nets using LoLA, a formal analysis tool.
-
A PostgreSQL database that stores structured information about patients, operations, pathways, tasks, and system users.
Communication between components is handled through secure HTTP (REST APIs), using JSON as the primary data format. Docker support is included to simplify deployment and ensure consistent runtime environments.
Technical Overview
- Frontend: React + Tailwind CSS
- Backend: Node.js + Express
- Microservices: Python + FastAPI
- Formal Analysis: LoLA for Petri net validation (checking liveness and absence of deadlocks)
- Database: PostgreSQL
- Deployment: Docker
Data Model
A key part of the system is its relational data model, designed to reflect the complexity of real-world clinical workflows. It ensures data consistency, supports extensibility, and provides a solid foundation for analysis.
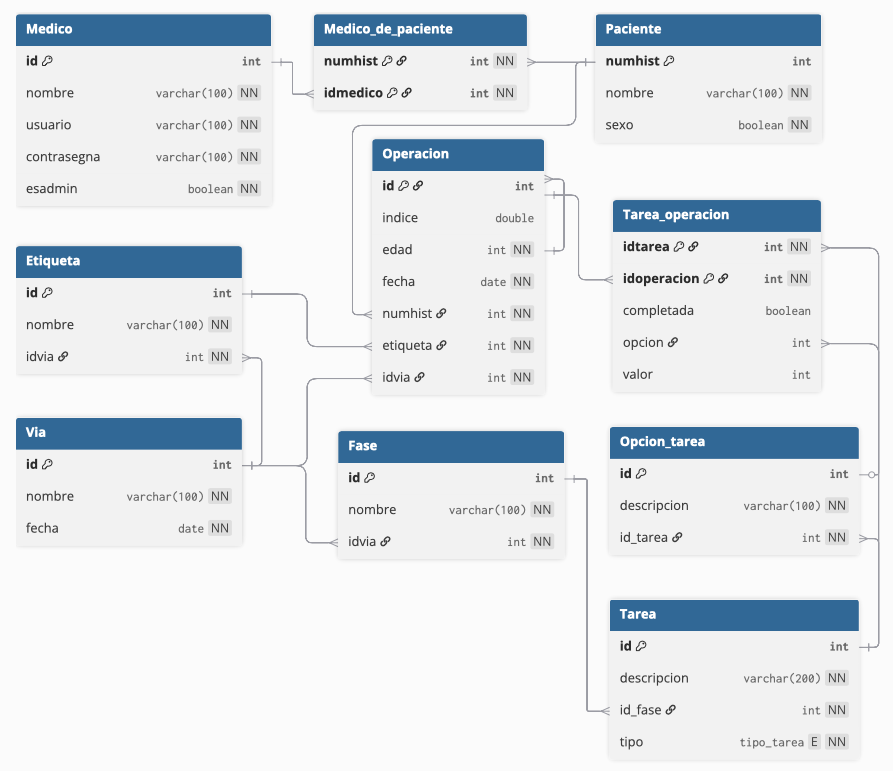
The main entities include:
- Patient: identified by medical record number, with personal and demographic information.
- Operation: a surgical procedure linked to a specific patient and clinical pathway.
- Clinical Pathway: a standardized treatment process, composed of multiple phases.
- Phase: a stage within the pathway, containing one or more tasks.
- Task: an action performed by the medical team, with defined response types (e.g., checkbox, numeric value, multiple choice).
- Option: possible responses to a task (for multiple-choice or binary inputs).
- User: hospital staff using the system, with roles (doctor or admin-doctor) determining permissions.
The database schema was iteratively refined using tools like DBML and dbdiagram.io to validate relationships and maintain normalization.
The Final Product
The result is a fully functional web platform that supports real clinical use. It provides two main user roles:
-
Doctors, who can:
- Search for patients by medical record number.
- View detailed information about past surgeries.
- Register a new operation using an existing clinical pathway.
- Track and document the completion of tasks during the procedure.
-
Admin-doctors, who have extended capabilities to:
- Import new pathway definitions from CSV files.
- Automatically generate a Petri net using the process mining algorithm.
- Validate the model using LoLA to ensure that it’s deadlock-free and live.
- Assign tasks to each phase and define expected responses.
- Edit and manage users, patients, and existing pathways.
The interface was designed to be clean, intuitive, and usable by non-technical medical staff, which was a consistent priority during the project. The backend includes audit-ready features and a complete API documented with OpenAPI (Swagger).
What’s Next?
While the current system is stable and technically validated, the next steps involve:
- User testing in a real hospital environment, with real patient data and surgical procedures.
- Gathering feedback from doctors and hospital IT teams to improve usability and reliability.
- Evaluating legal and ethical requirements for handling sensitive clinical data, including GDPR and hospital-specific protocols.
- Exploring deployment options, such as on-premise installations or hybrid cloud setups (e.g., database in-hospital, frontend on AWS).
The long-term vision is to expand the system into a generic platform for clinical process optimization, adaptable to other medical departments or even entire hospitals.
Challenges Faced
This project came with technical and organizational challenges:
- Integrating legacy code: The process mining algorithm was previously developed with minimal documentation. I had to reverse-engineer it and adapt it for dynamic input/output without rewriting it entirely.
- Choosing the right tools: Many Petri net analyzers (e.g., Renew, TimeNet) were not practical to integrate. I ultimately chose LoLA for its simplicity, Linux compatibility, and speed.
Lessons Learned
This project taught me much more than just how to build a full-stack application:
- Working with non-technical stakeholders is essential. Real progress happened when I translated clinical language into system logic and back again.
- Architecture matters early. Defining the system around microservices allowed me to scale, test, and debug more effectively than a monolithic approach would have.
- Documentation is part of the product. Tools like Swagger and Markdown-based API docs saved time and made integration more reliable.
- Iterative development works. Early demos and prototypes saved me from going too far in the wrong direction.
Above all, I learned how to design and deliver a usable, high-impact tool in a domain where precision and clarity are essential healthcare. This project took me through the entire software development lifecycle: from gathering real user requirements and prototyping, to designing architecture, implementing features, testing, and validating the final product. It taught me how software engineers work in practice balancing technical decisions with user needs, collaborating across disciplines, and building systems that are not just functional, but truly valuable in real-world settings. The result is a fully operational and validated platform that can improve how clinical pathways are managed from day one in a hospital environment.
You can find the complete source code and the documentation for this project on my GitHub repository: TFG
And if you’d like to see how the system works in action, check out the following video where I present the project and demonstrate its features:

